We use machine learning technology to do auto-translation. Click "English" on top navigation bar to check Chinese version.
Using Amazon Web Services Storage Gateway to modernize next-generation sequencing workflows
As we modernized our processes, the new solution needed to provide scalability and near real time to support rapid expansion or pop-up labs including the large volume of data transfer.
We needed management to reduce on-premises infrastructure/processing which takes too long. Speed was essential as we need to accelerate data migration to cloud to decrease turnaround time. Lastly, it was important that our solution could eliminate custom solutions for transfer and notification to increase operational efficiency through integration.
In this blog post, we will share our solution for real-time data transfer of lab data which was built using native Amazon Web Services technologies to scale and adapt to our increasing laboratory needs. Our solution uses
Solution overview
The NGS Data Lake is powered by Amazon Web Services Storage Gateway as the platform for data ingestion and notification, leveraging
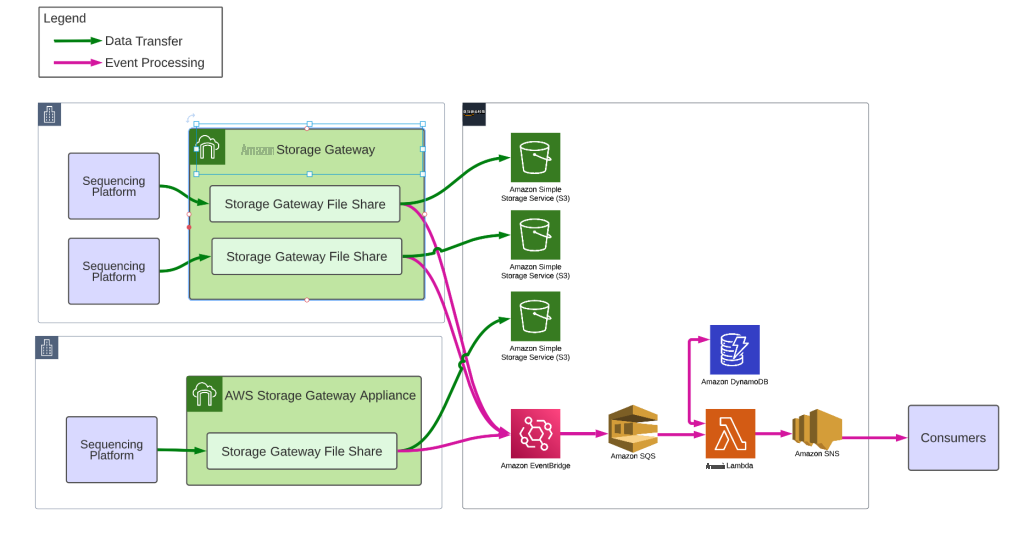
Amazon Web Services Storage Gateway
Event processing
Storage Gateway file shares emit
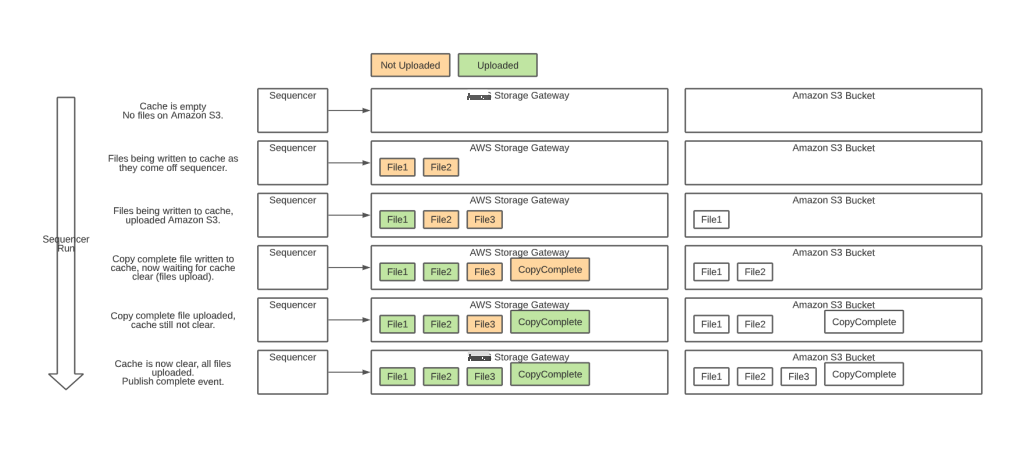
When the CopyComplete file appears, we confirm that all files in the run have been successfully uploaded to S3 by verifying that the v entire folder is empty. This straightforward step is powered by the Amazon Web Services Storage Gateway NotifyWhenUploaded API which sends an asynchronous confirmation when the cache is empty. When we receive this notification, we trigger a run complete event which flows through SNS to consumers. The data transfers in real time during the sequencing run, so our data uploads are often complete within minutes of the sequencing run finishing.
Data lake
In our data lake, each sequencing appliance has its own Amazon S3 bucket dedicated to the Amazon Web Services Storage Gateway file share. The entire data lake is provisioned through automation, so we can easily control bucket policies, lifecycle management, encryption, etc. We configure every bucket with inventories to a centralized bucket and appropriate replication and access logging policies. Our data lake is WORM (write once read many) so that our sequencing data is never modified or deleted. Consumers of the data lake are granted read only access per their requirements.
Deploying a new file share and Amazon S3 bucket to the data lake requires only updating a configuration document to place a new sequencer ID on an existing S3 File Gateway. The automation will provision a file share on the File Gateway and link it to a new Amazon S3 bucket using the shared data lake bucket configuration settings. All file shares and buckets are catalogued in a separate Amazon DynamoDB table during the automation process including relevant details on how to mount the file share, such as file share IP address and fileshare name. Because these resources are virtual, we can easily shift where file shares are deployed to move capacity around as needed. Onsite technical staff configure the sequencers to write to the file share once it has been provisioned and that concludes the installation process.
Deployment
There are two steps for deployment. We procure and install Amazon Web Services Storage Gateway devices if we don’t have enough existing capacity at the site. We have dashboards which show available capacity at each site, so that we know if we require additional hardware. If we require more hardware, we can order from our preferred reseller or if in the US, through
Conclusion
Exact Sciences has implemented Amazon Web Services Storage Gateway as the foundation for NGS Data Lake on Amazon Web Services, relying on the flexibility, scalability, ease of management, and native Amazon Web Services integrations with decoupled services to rapidly scale NGS data transfer solutions across the country. Since our initial deployment, we have uploaded hundreds of sequencing runs (many TB data) across 4 laboratory locations in 3 different time zones with a footprint of 9 Storage Gateway physical appliances serving 25 sequencing devices. Our infrastructure and provisioning process has become standardized and new sequencing platforms are brought online faster than ever. The solution requires a small capital investment, but scales indefinitely to provide robust processing for short term time-sensitive workloads and long-term data lake capabilities, all while lessening on-premises footprint in favor of relying on cloud native services.
The content and opinions in this post are those of the third-party author and Amazon Web Services is not responsible for the content or accuracy of this post.
The mentioned AWS GenAI Services service names relating to generative AI are only available or previewed in the Global Regions. Amazon Web Services China promotes AWS GenAI Services relating to generative AI solely for China-to-global business purposes and/or advanced technology introduction.